View on github
Current Tag
| Currently supports | |
|---|---|
| Binary-classification (currently using XGBoost and DNN) | Examples: DY vs ttbar, DY prompt vs DY fake, good electrons vs bad electrons |
| Multi-sample classification (currently using XGBoost and DNN) | Examples: DY vs (ttbar and QCD) |
| Multi-class classification (currently using XGBoost and DNN) | Examples: DY vs ttbar vs QCD, , good photons vs bad photons |
| Salient features: |
|---|
| Parallel reading of root files (using DASK) |
| Runs on flat ntuples (even NanoAODs) out of the box |
| Adding multiple MVAs is very trivial (Subject to available computing power) |
| Cross-section and pt-eta reweighting can be handled together |
| Multi-Sample training possible |
| Multi-Class training possible |
| Ability to customize thresholds |
| What will be the output of the trainer: |
|---|
| Feature distributions |
| Statistics in training and testing |
| ROCs, loss plots, MVA scores |
| Confusion Matrices |
| Correlation plots |
| Trained models (h5 (and pb) for DNN / pkl for XGBoost) |
| Optional outputs |
|---|
| 1) Threshold values of scores for chosen working points |
| 2) Efficiency vs pT and Efficiency vs eta plots for all classes |
| 3) Reweighting plots for pT and eta |
| 4) Comparison of new ID performance with benchmark ID flags |
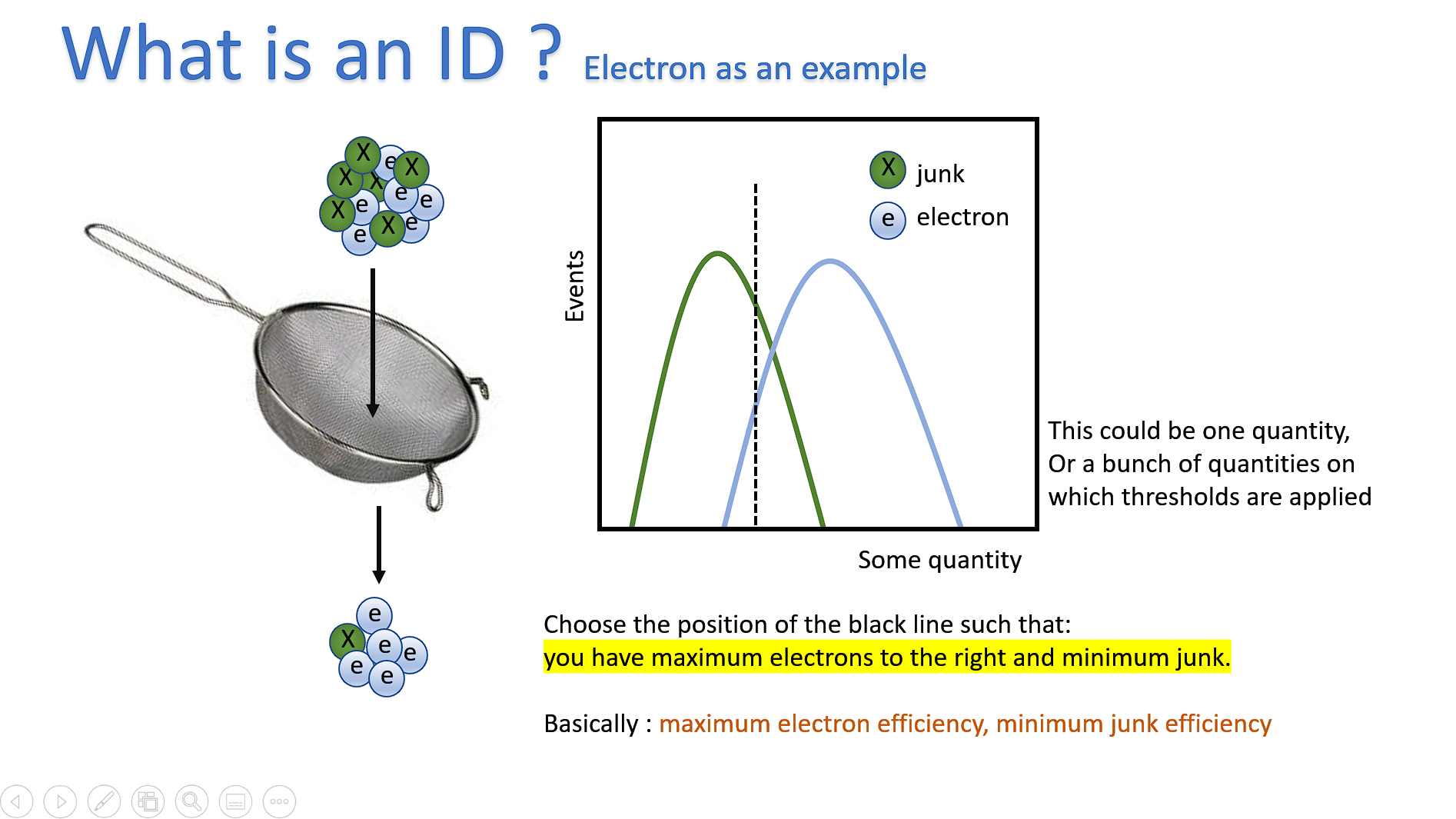
Primary intended use: For ID Training
Setting up
Clone using https
git clone -b v1.4.1 --depth 1 https://github.com/cms-egamma/ID-Trainer.git
Or
Clone using ssh
git clone -b v1.4.1 --depth 1 git@github.com:cms-egamma/ID-Trainer.git
Setup
In principle, you can set this up on your local computer by installing packages via conda/pip, but if possible please set up a cvmfs release.
When running on CPUs only
Use LCG 97python3 and you will have all the dependencies! (Tested at lxplus and SWAN)
source /cvmfs/sft.cern.ch/lcg/views/LCG_97python3/x86_64-centos7-gcc8-opt/setup.sh
When running with GPUs
The code can also transparently use a GPU, if a GPU card is available. Although, all packages need to be setup correctly.
For GPU in tensorflow, you can use the LCG_97py3cu10 cvmfs release:
source /cvmfs/sft.cern.ch/lcg/views/LCG_97py3cu10/x86_64-centos7-gcc7-opt/setup.sh
For XGBoost, while the code will use it automatically if you set UseGPU=True, it needs a GPU compiled XGBoost with CUDA >10.0. This is at the moment not possible with any cvmfs release.
You can certainly setup packages locally.
Running the trainer
Create a config
Create a new python config. Some sample python configs are available in the ‘Configs’ folder. They cover the most possible examples. All you need to do is to edit the config with the settings for your analysis and then run:
python Trainer.py NewTrainConfig #without the .py
The Trainer will read the settings from the config file and run training
Projects where the framework has been helpful
| 1) Run-3 Electron MVA ID |
| 2) Run-3 PF Electron ID |
| 3) Run-3 PF Photon ID |
| 4) Close photon analysis |
| 5) H->eeg analysis |
| 6) Resolved and merged ID |
##########################################
The different parts of the config
Imports
This is needed to use numpy and tensorflow. If you are using anything special within the config, you might have to add more imports.
import numpy as np
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Activation
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.layers import Dropout
from tensorflow.keras.callbacks import EarlyStopping
All the Parameters
| Parameters | Type | Description |
|---|---|---|
OutputDirName |
string | All plots, models, config file will be stored in this directory. This will be automatically created. If it already exists, it will overwrite everything if you run it again with the same OutputDirName |
branches |
list of strings | Branches to read (Should be in the input root files). Only these branches can be later used for any purpose. The ‘*’ is useful for selecting pattern-based branches. In principle one can do branches=["*"], but remember that the data loading time increases, if you select more branches |
SaveDataFrameCSV |
boolean | If True, this will save the data frame as a parquet file and the next time you run the same training with different parameters, it will be much faster |
loadfromsaved |
boolean | If root files and branches are the same as previous training and SaveDataFrameCSV was True, you can assign this as True, and data loading time will reduce significantly. Remember that this will use the same output directory as mentioned using OutputDirName, so the data frame should be present there |
Classes |
list of strings | Two or more classes possible. For two classes the code will do a binary classification. For more than two classes Can be anything but samples will be later loaded under this scheme. Example: Classes=['DY','TTBar'] or Classes=['Class1','Class2','Class3']. The order is important if you want to make an ID. In case of two classes, the first class has to be a Signal of interest. The second has to be a background. In multiclass, it does not matter which order one is using, but it is highly recommended that the first class is signal, if it is known. |
ClassColors |
list of strings | Colors for Classes to use in plots. Standard python colors work! |
Tree |
string | Location of the tree inside the root file |
processes |
list of process dictionaries | You can add as many process files as you like and assign them to a specific class. For example WZ.root and TTBar.root could be ‘Background’ class and DY.root could be ‘Signal’ or both ‘Signal and ‘background’ can come from the same root file. In fact you can have, as an example: 4 classes and 5 root files. The Trainer will take care of it at the backend. Look at the sample config below to see how processes are added. It is a list of process dictionaries. Each process has four options: Class, path, xsecwt,selection, with one example dictionary looking like this {'Class':'IsolatedSignal', 'path':['./DY.root','./Zee.root'], 'xsecwt': 1, 'selection':'(ele_pt > 5) & (abs(scl_eta) < 1.442) & (abs(scl_eta) < 2.5) & (matchedToGenEle==1)'}. At the end of the config adding processes in described in much detail. |
MVAs |
list of dictionaries | MVAs to use. You can add as many as you like: MVAtypes XGB and DNN are keywords, so names can be XGB_new, DNN_old etc, but keep XGB and DNN in the names, and no space please ( (That is how the framework identifies which algo to run). Look at the sample config below to see how MVAs are added. At the end of the config adding MVAs in described in much detail. |
Optional Parameters
| Parameters | Type | Description | Default value |
|---|---|---|---|
CMSLabel |
list of two strings | Left and Right titles for plots. Useful for adding things like “CMS Preliminary” and “13 TeV” on top of the plots. example: CMSLabel=["CMS Preliminary","13 TeV"] |
True |
Spectators |
list of features | Spectator variables which are plotted but not used for training | empty |
SpectatorBins |
list of binnings | Binning scheme for spectator variables which are plotted but not used for training | empty |
Reweighing |
boolean | This is independent of xsec reweighing (this reweighing will be done after taking into account xsec weight of multiple samples). Even if this is ‘False’, xsec reweighting will always be done. To switch off xsec reweighting, you can just assign the xsec weight is 1 |
False |
ptbins,etabins |
lists of numbers | $p_T$ and $\eta$ bins of interest (will be used for robustness studies: function coming soon) and will also be used for 2D $p_T$-$\eta$ reweighing if the Reweighing option is True |
Not activated until Reweighing==True |
ptwtvar,etawtvar |
strings | names of $p_T$ and $\eta$ branches | Not activated until Reweighing==True |
WhichClassToReweightTo |
string | 2D $p_T$-$\eta$ spectrum of all other classes will be reweighted to this class | Not activated until Reweighing==True |
OverlayWP |
list of strings | Working Point Flags to compare to (Should be in your ntuple and should also be read in branches) | empty list |
OverlayWPColors |
list of strings | Working Point Flags colors in plot (Usual hex-colors accepted) | empty list |
SigEffWPs |
list of strings | To print thresholds of mva scores for corresponding signal efficiency, example ["80%","90%"] (Add as many as you would like) |
empty list |
testsize |
float | In fraction, how much data to use for testing (0.3 means 30%) | 0.2 |
flatten |
boolean | For NanoAOD and other un-flattened trees, you can assign this as True to flatten branches with variable length for each event (Event level -> Object level) |
False |
Debug |
boolean | If True, only a small subset of events/objects are used for either Signal or background. Useful for quick debugging of code | False |
RandomState |
integer | Choose the same number every time for reproducibility | 42 |
MVAlogplot |
boolean | If true, MVA outputs are plotted in log scale | False |
ROClogplot |
boolean | If true, ROC is plotted in log scale | False |
Multicore |
boolean | If True all CPU cores available are used XGB | True |
How to add more variables (that are a not directly in the trees)? or modify the ones that are in tree
| Function | Type | Description | Default value |
|---|---|---|---|
modifydf |
function | In your config, you can add a function with this exact name modifydf which accepts a pandas dataframe and manipulates it and then returns the dataframe. |
Not activated until defined |
Using this you can add new variables or modify already present variables. A very simple example: def modifydf(df): df['A']=df[X]+df[Y]; return df; This will add a new branch named ‘A’.
There are lot of things you can do with a modifydf.
def modifydf(df):
#Example: Simple additions
df["sumleptonpT"]=df["muonpt"]+df["electronpt"]
#Example: Check if all selections are passed and store as new variables
for ind, event in df.iterrows():
print(f'Event {ind}')
for Muonpt, Muoneta in zip(event.Muon_pt,event.Muon_eta):
#print(f'Muons: {Muonpt} and {Muoneta}')
if (Muonpt>26) and (abs(Muoneta)<2.1):
print(f'Muons passed')
df.loc[ind,'MuonBit']=1
break
for Jetpt, Jeteta in zip(event.Jet_pt,event.Jet_eta):
#print(f'Jets: {Jetpt} and {Jeteta}')
if (Jetpt>30) and (abs(Jeteta)<2.4):
print(f'jets passed')
df.loc[ind,'JetBit']=1
break
for JetpfCombinedInclusiveSecondaryVertexV2BJetTags in event.Jet_pfCombinedInclusiveSecondaryVertexV2BJetTags:
if JetpfCombinedInclusiveSecondaryVertexV2BJetTags>0.627:
print(f'pf also passed')
df.loc[ind,'pfBit']=1
break
continue
#Example: Loop over gen particles
df['GenToppt']=-999
df['GenTopeta']=-999
for ind, row in df.iterrows():
#print(f'Top pTs for Event {ind}')
for partind, part in enumerate(row.Gen_pdg_id):
if abs(row.Gen_pdg_id[partind])==6 and row.Gen_numDaught[partind]==2:
#print(f'Found top at {partind} with pT {row.Gen_pt[partind]} GeV and mother ID {row.Gen_motherpdg_id[partind]} with {row.Gen_numDaught[partind]} daughters')
df.loc[ind,'GenToppt'] = row.Gen_pt[partind]
df.loc[ind,'GenTopeta'] = row.Gen_eta[partind]
return df
A sample config for running XGboost and DNN together
#####################################################################
######Do not touch this
import numpy as np
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Activation
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.layers import Dropout
from tensorflow.keras.callbacks import EarlyStopping
#####################################################################
####Start here
#####################################################################
OutputDirName = 'SimpleBinaryClassification' #All plots, models, config file will be stored here
Debug=False # If True, only a small subset of events/objects are used for either Signal or background #Useful for quick debugging
#Branches to read #Should be in the root files #Only the read branches can be later used for any purpose
branches=["scl_eta","ele*","matched*","EleMVACats",'passElectronSelection','Fall*']
SaveDataFrameCSV,loadfromsaved=True,False #If loadfromsaved=True, dataframe stored in OutputDirName will be read
Classes,ClassColors = ['IsolatedSignal','NonIsolated'],['#377eb8', '#ff7f00']
#Remeber: For binary classification, first class of the Classes argument should be signal, otherwise, plots might not make sense.
processes=[
{'Class':'IsolatedSignal','path':['./DY.root','./Zee.root'],
#Can be a single root file, a list of root file, or even a folder but in a tuple format (folder,fileextension), like ('./samples','.root')
'xsecwt': 1, #can be a number or a branch name, like 'weight' #Will go into training
'selection':'(ele_pt > 5) & (abs(scl_eta) < 1.442) & (abs(scl_eta) < 2.5) & (matchedToGenEle==1)', #selection
},
{'Class':'NonIsolated','path':['./QCD.root'],
#Can be a single root file, a list of root file, or even a folder but in a tuple format (folder,fileextension), like ('./samples','.root')
'xsecwt': 1, #can be a number or a branch name, like 'weight' #Will go into training
'selection':'(ele_pt > 5) & (abs(scl_eta) < 1.442) & (abs(scl_eta) < 2.5) & (matchedToGenEle==0)', #selection
},
]
Tree = "ntuplizer/tree"
#MVAs to use as a list of dictionaries
MVAs = [
#can add as many as you like: For MVAtypes XGB and DNN are keywords, so names can be XGB_new, DNN_old etc.
#But keep XGB and DNN in the names (That is how the framework identifies which algo to run
{"MVAtype":"XGB_1", #Keyword to identify MVA method.
"Color":"green", #Plot color for MVA
"Label":"XGB try1", # label can be anything (this is how you will identify them on plot legends)
"features":["ele_fbrem", "ele_deltaetain", "ele_deltaphiin", "ele_oldsigmaietaieta",
"ele_oldhe", "ele_ep", "ele_olde15", "ele_eelepout"],
"feature_bins":[100 , 100, 100, 100, 100, 100, 100, 100], #same length as features
#Binning used only for plotting features (should be in the same order as features), does not affect training
'Scaler':"MinMaxScaler", #Scaling for features before passing to the model training
'UseGPU':True, #If you have a GPU card, you can turn on this option (CUDA 10.0, Compute Capability 3.5 required)
"XGBGridSearch":{'min_child_weight': [5], 'max_depth': [2,3,4]} ## multiple values for a parameter will automatically do a grid search
#All standard XGB parameters supported
},
{"MVAtype":"DNN_clusteriso_2drwt",#Keyword to identify MVA method.
"Color":"black", #Plot color for MVA
"Label":"DNN_clusteriso_2drwt", # label can be anything (this is how you will identify them on plot legends)
"features":["ele_fbrem", "ele_deltaetain", "ele_deltaphiin", "ele_oldsigmaietaieta",
"ele_oldhe", "ele_ep", "ele_olde15", "ele_eelepout",
"ele_kfchi2", "ele_kfhits", "ele_expected_inner_hits","ele_dr03TkSumPt",
"ele_dr03EcalRecHitSumEt","ele_dr03HcalTowerSumEt","ele_gsfchi2","scl_eta","ele_pt",
'ele_nbrem','ele_deltaetaseed','ele_hadronicOverEm','ele_olde25max','ele_olde55'],
"feature_bins":[100 for i in range(22)], #same length as features
'Scaler':"MinMaxScaler", #Scaling for features before passing to the model training
"DNNDict":{'epochs':10, 'batchsize':5000,
'model': Sequential([Dense(24, kernel_initializer='glorot_normal', activation='relu'),
Dense(48, activation="relu"),
Dense(24, activation="relu"),
Dropout(0.1),
Dense(len(Classes),activation="softmax")]),
'compile':{'loss':'categorical_crossentropy','optimizer':Adam(lr=0.001), 'metrics':['accuracy']},
'earlyStopping': EarlyStopping(monitor='val_loss', mode='min', verbose=1, patience=10)
}
},
]
More on how to add processes and MVAs along with neat functionalities
Processes
If you add a dictionary like this in process, it will add the selected branches from the root files and assign them to specified class.
{'Class':'IsolatedSignal','path':['./DY.root','./Zee.root'],
'xsecwt': 1, #can be a number or a branch name, like 'weight' #Will go into training
'selection':'(ele_pt > 5) & (abs(scl_eta) < 1.442) & (abs(scl_eta) < 2.5) & (matchedToGenEle==1)', #selection
}
The Class argument directly links to the ‘Classes’ argument in the config file.
The path argument can be a single root file, a list of root files, or even a folder but in a tuple format (folder,fileextension), like (‘./samples’,’.root’)
||
|—|
|example:'path':['./DY.root','./Zee.root']|
|example:'path':'./DY.root'|
|example:'path':('/samples/','.root')|
The xsecwt argument can be number, branch name or even a branch name with a multiplier factor as well.
||
|—|
|example: 'xsecwt': 1 |
|example: 'xsecwt': "weight" |
|example: 'xsecwt': ("weight",5)|
The selection argument will take & and | for and and or. You can place selection on branches in the tree. Remember what selection can you place will depend on what branches are in the tree.
MVAs
MVAs to use. You can add as many as you like: MVAtypes XGB and DNN are keywords, so names can be XGB_new, DNN_old etc, but keep XGB and DNN in the names, and no space please ( (That is how the framework identifies which algo to run). Look at the sample config below to see how MVAs are added. At the end of the config adding MVAs in described in much detail.
MVAs = [
#can add as many as you like: For MVAtypes XGB and DNN are keywords, so names can be XGB_new, DNN_old etc.
#But keep XGB and DNN in the names (That is how the framework identifies which algo to run
{"MVAtype":"XGB_1", #Keyword to identify MVA method.
"Color":"green", #Plot color for MVA
"Label":"XGB try1", # label can be anything (this is how you will identify them on plot legends)
"features":["ele_fbrem", "ele_deltaetain", "ele_deltaphiin", "ele_oldsigmaietaieta",
"ele_oldhe", "ele_ep", "ele_olde15", "ele_eelepout"],
"feature_bins":[100 , 100, 100, 100, 100, 100, 100, 100], #same length as features
#Binning used only for plotting features (should be in the same order as features), does not affect training
'Scaler':"MinMaxScaler", #Scaling for features before passing to the model training
'UseGPU':True, #If you have a GPU card, you can turn on this option (CUDA 10.0, Compute Capability 3.5 required)
"XGBGridSearch":{'min_child_weight': [5], 'max_depth': [2,3,4]} ## multiple values for a parameter will automatically do a grid search
#All standard XGB parameters supported
},
{"MVAtype":"DNN_clusteriso_2drwt",#Keyword to identify MVA method.
"Color":"black", #Plot color for MVA
"Label":"DNN_clusteriso_2drwt", # label can be anything (this is how you will identify them on plot legends)
"features":["ele_fbrem", "ele_deltaetain", "ele_deltaphiin", "ele_oldsigmaietaieta",
"ele_oldhe", "ele_ep", "ele_olde15", "ele_eelepout",
"ele_kfchi2", "ele_kfhits", "ele_expected_inner_hits","ele_dr03TkSumPt",
"ele_dr03EcalRecHitSumEt","ele_dr03HcalTowerSumEt","ele_gsfchi2","scl_eta","ele_pt",
'ele_nbrem','ele_deltaetaseed','ele_hadronicOverEm','ele_olde25max','ele_olde55'],
"feature_bins":[100 for i in range(22)], #same length as features
'Scaler':"MinMaxScaler", #Scaling for features before passing to the model training
"DNNDict":{'epochs':10, 'batchsize':100,
'model': Sequential([Dense(24, kernel_initializer='glorot_normal', activation='relu'),
Dense(48, activation="relu"),
Dense(24, activation="relu"),
Dropout(0.1),
Dense(len(Classes),activation="softmax")]),
'compile':{'loss':'categorical_crossentropy','optimizer':Adam(lr=0.001), 'metrics':['accuracy']},
'earlyStopping': EarlyStopping(monitor='val_loss', mode='min', verbose=1, patience=10)
}
},
]



